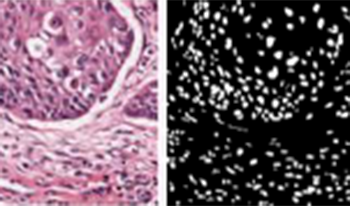
This paper introduces a novel framework for generating high-quality images from “visual sentences” extracted from video sequences. By combining a lightweight autoregressive model with a Vector Quantized Generative Adversarial Network (VQGAN), our approach achieves a favorable trade-off between computational efficiency and image fidelity. Unlike conventional methods that require substantial resources, the proposed framework efficiently captures sequential patterns in partially annotated frames and synthesizes coherent, contextually accurate images. Empirical results demonstrate that our method not only attains state-of-the-art performance on various benchmarks but also reduces inference overhead, making it well-suited for real-time and resource-constrained environments. Furthermore, we explore its applicability to medical image analysis, showcasing robust denoising, brightness adjustment, and segmentation capabilities. Overall, our contributions highlight an effective balance between performance and efficiency, paving the way for scalable and adaptive image generation across diverse multimedia domains.

In medical segmentation, the acquisition of high-quality labeled data remains a significant challenge due to the substantial cost and time required for expert annotations. Variability in imaging conditions, patient diversity, and the use of different imaging devices further complicate model training. The high dimensionality of medical images also imposes considerable computational demands, while small lesions or abnormalities can create class imbalance, hindering segmentation accuracy. Pre-training on synthetic datasets in medical imaging may enable Vision Transformers (ViTs) to develop robust feature representations, even during the fine-tuning phase, when high-quality labeled data is limited. In this work, we propose integrating Formula-Driven Supervised Learning (FDSL) synthetic datasets with medical imaging to enhance pre-training for segmentation tasks. We implemented a custom fractal dataset, Style Fractals, capable of generating high-resolution images, including those measuring 8k x 8k pixels. Our results indicate improved performance when using the SAM model for segmentation, in conjunction with robust augmentation techniques, increasing performance from 62.30% to 63.68%. This was followed by fine-tuning on the PAIP dataset, a high-resolution, real-world pathology dataset focused on liver cancer. Additionally, we present results using another synthetic dataset, SegRCDB, for comparative analysis.
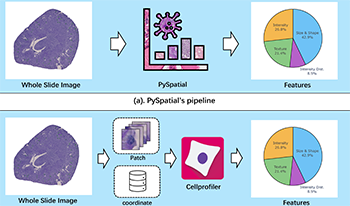
Whole Slide Image (WSI) analysis plays a crucial role in modern digital pathology, enabling large-scale feature extraction from tissue samples. However, traditional feature extraction pipelines based on tools like CellProfiler often involve lengthy workflows, requiring WSI segmentation into patches, feature extraction at the patch level, and subsequent mapping back to the original WSI. To address these challenges, we present PySpatial, a high-speed pathomics toolkit specifically designed for WSI-level analysis. PySpatial streamlines the conventional pipeline by directly operating on computational regions of interest, reducing redundant processing steps. Utilizing rtreebased spatial indexing and matrix-based computation, PySpatial efficiently maps and processes computational regions, significantly accelerating feature extraction while maintaining high accuracy. Our experiments on two datasets—Perivascular Epithelioid Cell (PEC) and data from the Kidney Precision Medicine Project (KPMP)—demonstrate substantial performance improvements. For smaller and sparse objects in PEC datasets, PySpatial achieves nearly a 10-fold speedup compared to standard CellProfiler pipelines. For larger objects, such as glomeruli and arteries in KPMP datasets, PySpatial achieves a 2-fold speedup. These results highlight PySpatial’s potential to handle large-scale WSI analysis with enhanced efficiency and accuracy, paving the way for broader applications in digital pathology.
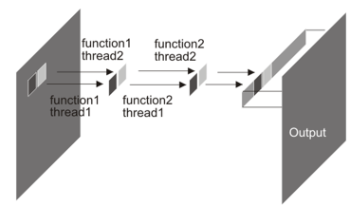
libvips is a LGPL licensed (open source and free for commercial use), portable, horizontally-threaded, demand-driven, 2D image processing library with its origins in imaging research projects. Compared to similar libraries, libvips runs quickly and uses little memory. It supports numeric formats from 8-bit integer to 128-bit complex, any number of color separation bands, most popular image formats, and many specialized scientific image formats. It has become popular in applications such as virtual microscopy and art imaging, and very popular as an image processing library for the web. This paper outlines the history of the library, explains how libvips achieves its good performance, presents benchmarks, and gives an overview of the implementation and of the wider libvips ecosystem.

This study evaluates the benefit of using parallelism from GPUs or multi-core CPUs for particle advection workloads. We perform 1000+ experiments, involving four generations of Nvidia GPUs, four CPUs with varying numbers of cores, two particle advection algorithms, many different workloads (i.e., number of particles and number of steps), and, for GPU tests, performance with and without data transfer. The results inform whether or not a visualization developer should incorporate parallelism in their code, what type (CPU or GPU), and the key factors influencing performance. Finally, we find that CPU parallelism is the better choice for most common workloads, even when ignoring costs for data transfer.
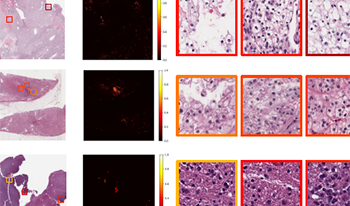
Multi-modal learning adeptly integrates visual and textual data, but its application to histopathology image and text analysis remains challenging, particularly with large, high-resolution images like gigapixel Whole Slide Images (WSIs). Current methods typically rely on manual region labeling or multi-stage learning to assemble local representations (e.g., patch-level) into global features (e.g., slide-level). However, there is no effective way to integrate multi-scale image representations with text data in a seamless end-to-end process. In this study, we introduce Multi-Level Text-Guided Representation End-to-End Learning (mTREE). This novel text-guided approach effectively captures multi-scale WSI representations by utilizing information from accompanying textual pathology information. mTREE innovatively combines – the localization of key areas (“global-tolocal”) and the development of a WSI-level image-text representation (“local-to-global”) – into a unified, end-to-end learning framework. In this model, textual information serves a dual purpose: firstly, functioning as an attention map to accurately identify key areas, and secondly, acting as a conduit for integrating textual features into the comprehensive representation of the image. Our study demonstrates the effectiveness of mTREE through quantitative analyses in two image-related tasks: classification and survival prediction, showcasing its remarkable superiority over baselines. Code and trained models are made available at https://github.com/hrlblab/mTREE.
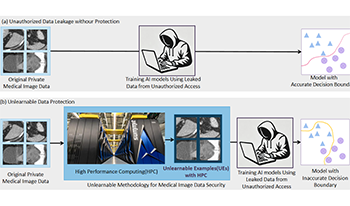
Recent advancements in AI models, like ChatGPT, are structured to retain user interactions, which could inadvertently include sensitive healthcare data. In the healthcare field, particularly when radiologists use AI-driven diagnostic tools hosted on online platforms, there is a risk that medical imaging data may be repurposed for future AI training without explicit consent, spotlighting critical privacy and intellectual property concerns around healthcare data usage. Addressing these privacy challenges, a novel approach known as Unlearnable Examples (UEs) has been introduced, aiming to make data unlearnable to deep learning models. A prominent method within this area, called Unlearnable Clustering (UC), has shown improved UE performance with larger batch sizes but was previously limited by computational resources (e.g., a single workstation). To push the boundaries of UE performance with theoretically unlimited resources, we scaled up UC learning across various datasets using Distributed Data Parallel (DDP) training on the Summit supercomputer. Our goal was to examine UE efficacy at high-performance computing (HPC) levels to prevent unauthorized learning and enhance data security, particularly exploring the impact of batch size on UE’s unlearnability. Utilizing the robust computational capabilities of the Summit, extensive experiments were conducted on diverse datasets such as Pets, MedMNist, Flowers, and Flowers102. Our findings reveal that both overly large and overly small batch sizes can lead to performance instability and affect accuracy. However, the relationship between batch size and unlearnability varied across datasets, highlighting the necessity for tailored batch size strategies to achieve optimal data protection. The use of Summit’s high-performance GPUs, along with the efficiency of the DDP framework, facilitated rapid updates of model parameters and consistent training across nodes. Our results underscore the critical role of selecting appropriate batch sizes based on the specific characteristics of each dataset to prevent learning and ensure data security in deep learning applications. The source code is publicly available at https: // github. com/ hrlblab/ UE_ HPC .
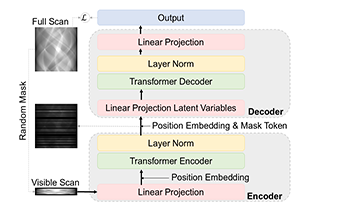
Sinogram inpainting is a critical task in computed tomography (CT) imaging, where missing or incomplete sinograms can significantly decrease image reconstruction quality. High-quality sinogram inpainting is essential for achieving high-quality CT images, enabling better diagnosis and treatment. To address this challenge, we propose SinoTx, a model based on the Transformer architecture specifically designed for sinogram completion. SinoTx leverages the inherent strengths of Transformers in capturing global dependencies, making it well-suited for handling the complex patterns present in sinograms. Our experimental results demonstrate that SinoTx outperforms existing baseline methods, achieving up to a 32.3% improvement in the Structural Similarity Index (SSIM) and a 44.2% increase in Peak Signal-to-Noise Ratio (PSNR).