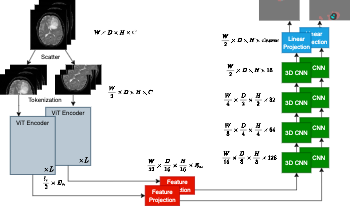
We introduce an efficient distributed sequence parallel approach for training transformer-based deep learning image segmentation models. The neural network models are comprised of a combination of a Vision Transformer encoder with a convolutional decoder to provide image segmentation mappings. The utility of the distributed sequence parallel approach is especially useful in cases where the tokenized embedding representation of image data are too large to fit into standard computing hardware memory. To demonstrate the performance and characteristics of our models trained in sequence parallel fashion compared to standard models, we evaluate our approach using a 3D MRI brain tumor segmentation dataset. We show that training with a sequence parallel approach can match standard sequential model training in terms of convergence. Furthermore, we show that our sequence parallel approach has the capability to support training of models that would not be possible on standard computing resources.

We introduce a physics guided data-driven method for image-based multi-material decomposition for dual-energy computed tomography (CT) scans. The method is demonstrated for CT scans of virtual human phantoms containing more than two types of tissues. The method is a physics-driven supervised learning technique. We take advantage of the mass attenuation coefficient of dense materials compared to that of muscle tissues to perform a preliminary extraction of the dense material from the images using unsupervised methods. We then perform supervised deep learning on the images processed by the extracted dense material to obtain the final multi-material tissue map. The method is demonstrated on simulated breast models with calcifications as the dense material placed amongst the muscle tissues. The physics-guided machine learning method accurately decomposes the various tissues from input images, achieving a normalized root-mean-squared error of 2.75%.