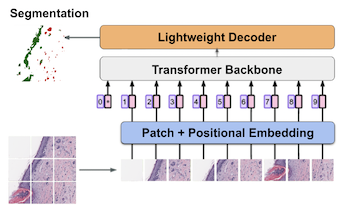
Prognosis for melanoma patients is traditionally determined with a tumor depth measurement called Breslow thickness. However, Breslow thickness fails to account for cross-sectional area, which is more useful for prognosis. We propose to use segmentation methods to estimate cross-sectional area of invasive melanoma in whole-slide images. First, we design a custom segmentation model from a transformer pretrained on breast cancer images, and adapt it for melanoma segmentation. Secondly, we finetune a segmentation backbone pretrained on natural images. Our proposed models produce quantitatively superior results compared to previous approaches and qualitatively better results as verified through a dermatologist.
Regression-based radiance field reconstruction strategies, such as neural radiance fields (NeRFs) and, physics-based, 3D Gaussian splatting (3DGS), have gained popularity in novel view synthesis and scene representation. These methods parameterize a high-dimensional function that represents a radiance field, from a low-dimensional camera input. However, these problems are ill-posed and struggle to represent high (spatial) frequency data; manifesting as reconstruction artifacts when estimating high frequency details such as small hairs, fibers, or reflective surfaces. Here we show that classical spherical sampling around a target, often referred to as sampling a bounded scene, inhomogeneously samples the targets Fourier domain, resulting in spectral bias in the collected samples. We generalize the ill-posed problems of view-synthesis and scene representation as expressions of projection tomograpy and explore the upper-bound reconstruction limits of regression-based and integration-based strategies. We introduce a physics-based sampling strategy that we directly apply to 3DGS, and demonstrate high fidelity 3D anisotropic radiance field reconstructions with reconstruction PSNR scores as high as 44.04 dB and SSIM scores of 0.99, following the same metric analysis as defined in Mip-NeRF360.
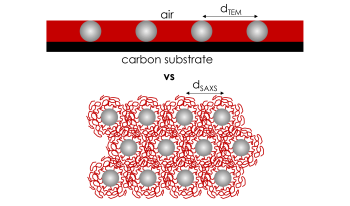
In the field of polymers, 2D images are often used to discern information about the microstructure of bulk polymer materials. For brush particle assembly structures, this work evaluates microstructure information retrieved from different material characterization techniques for thin film (i.e., electron imaging of brush particle monolayers) and bulk materials (small angle X-ray scattering), respectively. The effect of confinement of polymer chains into thin (2D) films on the conformation of tethered chains is discussed and used to rationalize systematic discrepancies between characteristic nanoparticle spacings in thin films and bulk materials. An approach to rationalize bulk material properties based on thin film measurements is presented.
Traditional super-resolution processing computes sub-pixel alignment over a sequence of image captures to allow sampling at a finer spatial resolution. Alternatively, the mechanism intended to implement in-body image stabilization (IBIS) can be used to shift the sensor in a stationary camera by precise fractions of a pixel between exposures. The implicitly perfect alignment of pixel-shift images reduces post-processing to interleaving of raw data, but motion of camera or scene elements produces disturbing artifacts. Determining misalignments on raw images from cameras using color filter array (CFA) sensors is potentially problematic, so the synthesized super-resolution image is instead typically built from already-interpolated image data, with a reduction in tonal quality. The current work instead directly models the certainty, or confidence, with which pixel values are known. Sub-pixel alignment may be computed on either raw or interpolated image data. Still, only the underlying raw samples have precisely known values, so only they are used to compute the super-resolution image. However, primarily due to motion, even raw pixel values can have variable value certainty. Thus, a confidence metric is calculated for each raw pixel value and used as a weighting factor in computing the best estimate for the value of each super-resolution pixel.

Acquisitions of mass-per-charge (m/z) spectrometry data from tissue samples, at high spatial resolutions, using Mass Spectrometry Imaging (MSI), require hours to days of time. The Deep Learning Approach for Dynamic Sampling (DLADS) and Supervised Learning Approach for Dynamic Sampling with Least-Squares (SLADS-LS) algorithms follow compressed sensing principles to minimize the number of physical measurements performed, generating low-error reconstructions from spatially sparse data. Measurement locations are actively determined during scanning, according to which are estimated, by a machine learning model, to provide the most relevant information to an intended reconstruction process. Preliminary results for DLADS and SLADS-LS simulations with Matrix-Assisted Laser Desorption/Ionization (MALDI) MSI match prior 70% throughput improvements, achieved in nanoscale Desorption Electro-Spray Ionization (nano-DESI) MSI. A new multimodal DLADS variant incorporates optical imaging for a 5% improvement to final reconstruction quality, with DLADS holding a 4% advantage over SLADS-LS regression performance. Further, a Forward Feature Selection (FFS) algorithm replaces expert-based determination of m/z channels targeted during scans, with negligible impact to location selection and reconstruction quality.

The dynamic range of the intensity of long-wave infrared (LWIR) cameras are often more than 8bit and its images have to be visualized using histogram equalization and so on. Many visualization methods do not consider effects of noise, which must be taken care of in real situations. We propose a novel LWIR images visualization method based on gradient-domain processing or gradient mapping. Processing based on intensity and gradient power in the gradient domain enables visualizing LWIR images with noise reduction. We evaluate the proposed method quantitatively and qualitatively and show its effectiveness.
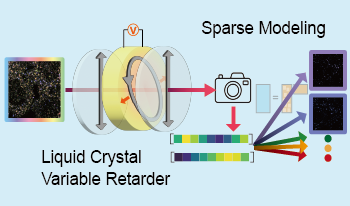
In spatial transcriptomics, which allows the analysis of gene expression while preserving its location in tissues, RNA molecules are hybridized with a fluorescent-labeled DNA probe for detection. In this study, we aim to improve the efficiency of spatial transcriptomics by simultaneously using multiple fluorescent dyes with overlapping spectra. We propose a method to quantify each fluorescent dyes using a liquid crystal variable retarder as a spectral modulator, which can control the spectral transmittance by changing the voltage. The spectrum of light passing through the modulator is integrated by the image sensor and observed as intensity. We quantify the fluorescent dyes at each pixel using intensities of various spectral transmittances as a spectral code and applying sparse modeling using a dictionary created by simulating observations for the fluorescent dyes used in hybridization. We verified the principle of the proposed method and demonstrated its feasibility through simulation experiments.
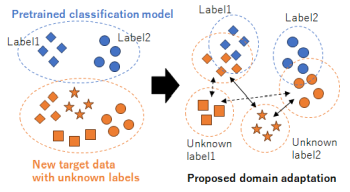
Domain adaptation, which transfers an existing system with teacher labels (source domain) to another system without teacher labels (target domain), has garnered significant interest to reduce human annotations and build AI models efficiently. Open set domain adaptation considers unknown labels in the target domain that were not present in the source domain. Conventional methods treat unknown labels as a single entity, but this assumption may not hold true in real-world scenarios. To address this challenge, we propose open set domain adaptation for image classification with multiple unknown labels. Assuming that there exists a discrepancy in the feature space between the known labels in the source domain and the unknown labels in the target domain based on their type, we can leverage clustering to classify the types of unknown labels by considering the pixel-wise feature distances between samples in the target domain and the known labels in the source domain. This enables us to assign pseudo-labels to target samples based on the classification results obtained through unsupervised clustering with an unknown number of clusters. Experimental results show that the accuracy of domain adaptation is improved by re-training using these pseudo-labels in a closed set domain adaptation setting.