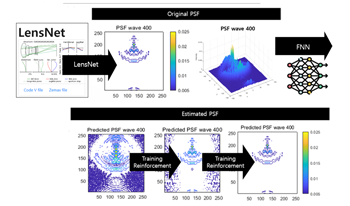
The initiation of CMOS simulations involves input scene data composed of wavelength-specific radiance, for which the application of lens PSF effects necessitates monochromatic PSFs corresponding to each wavelength. However, most lens manufacturers supply clients with lens files that contain polychromatic PSFs. Typically, upon opening these lens design files, only the weights for each wavelength can be discerned. Therefore, the ability to decompose these polychromatic PSFs into their monochromatic constituents is essential to accurately apply lens effects in CMOS simulations and to facilitate the intricate analysis of light interactions within optical systems. To address this need, our study utilizes deep learning techniques to decompose polychromatic PSFs into their constituent monochromatic elements. Leveraging lens data obtained from LensNet, we construct a polychromatic PSF which serves as the input for our deep neural network model. This model is specially trained to predict the monochromatic PSFs, revealing the distinct characteristics of each wavelength involved. The effectiveness of our approach is validated through extensive testing and detailed visualization methods. These include both 2D contour plots and 3D surface plots, which confirm the model's capability to accurately extract the monochromatic PSFs. This process is not only vital for current optical analysis but also paves the way for future advancements in neural network architectures and machine learning methodologies to refine the extraction process.

In this paper, we introduce silhouette tomography, a novel formulation of X-ray computed tomography that relies only on the geometry of the imaging system. We formulate silhouette tomography mathematically and provide a simple method for obtaining a particular solution to the problem, assuming that any solution exists. We then propose a supervised reconstruction approach that uses a deep neural network to solve the silhouette tomography problem. We present experimental results on a synthetic dataset that demonstrate the effectiveness of the proposed method.
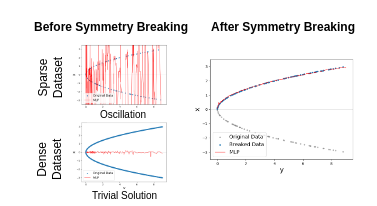
For nonlinear inverse problems that are prevalent in imaging science, symmetries in the forward model are common. When data-driven deep learning approaches are used to solve such problems, such intrinsic symmetries can cause substantial learning difficulties. In this paper, we explain how such difficulties arise and, more importantly, how to overcome them by preprocessing the training set before any learning, i.e., symmetry breaking. We take the far-field Fourier phase retrieval, which is central to many areas of scientific imaging, as an example and show that symmetric breaking can substantially improve data-driven learning performance. We also formulate the principle of symmetry breaking that can lead to efficient learning.
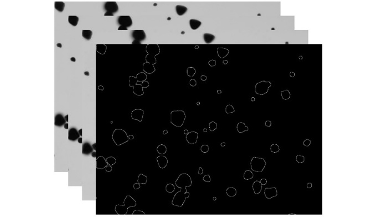
Starch plays a pivotal role in human society, serving as a vital component of our food sources and finding widespread applications in various industries. Microscopic imaging offers a straightforward, efficient, and precise approach to examine the distribution, morphology, and dimensions of starch granules. Quantitative analysis through the segmentation of starch granules from the background aids researchers in exploring their physicochemical properties. This article presents a novel approach utilizing a modified U-Net model in deep learning to achieve the segmentation of starch granule microscope images with remarkable accuracy. The method yields impressive results, with mean values for several evaluation metrics including JS, Dice, Accuracy, Precision, Sensitivity and Specificityreaching 89.67%, 94.55%, 99.40%, 94.89%, 94.23% and 99.70%, respectively.